Day :
Session Introduction
James Lyons-Weiler
Institute for Pure and Applied Knowledge,USA
Title: Personalized Immunity: Lose the Bias, Secure the Public Trust

Biography:
Lyons-Weiler is the CEO of The Institute for Pure and Applied Knowledge. He has published over 50 peer-reviewed studies in well-respected journals, has play a key role in over 100 basic, clinical and translational research studies, and has written three books on biomedicine and biomedical research. Dr. Lyons-Weiler is expert in translational research, genomics, genetics, proteomics, biomarkers, systems biology, and conducts research on cellular and molecular responses to traumatic brain injury and the neuroimmunological responses to environmental toxins. Scientists at IPAK focus on reducing human pain and suffering through knowledge.
Abstract:
As translational research goes, vaccine safety science has a deplorable record that now places the artificial immunization enterprise at real risk. Examples of research practices that border on fraud and certainly fit an agenda of risk perception minimization are well-known to the public, and this knowledge translates into vaccine hesitancy and refusal. Pushes for mandates without exemptions alienate the public to both medicine and science. Truly objective vaccine risk management via personalized immunity and respect for informed consent and choice will help practitioners build better relationships with their patients. A re-assessment of strategies to artificial immunization is needed. To that end, seven tracts, founded on thousands of research studies, are outlined that will help bring artificial immunization into the 21st Century: (1) reformulation, (2) risk indicators and biomarkers, (3) scheduling, (4) respect for laws and regulations governing informed consent, (5) regulating conflicts of interest, (6) regulatory reform, and (7) enforcement of adverse event reporting are all areas ripe for improvement. An era of cottage-industry innovation in artificial immunization competing on the platform of safety is needed to foster competition.

Biography:
Sergey Suchkov graduated from Astrakhan State Medical University with MD, then in 1985 maintained his PhD at I.M.Sechenov Moscow Medical Academy and in 2001, his Doctorship at the Nat Inst of Immunol, Russia. From 1987 through 1989, worked for Koltzov Inst of Dev Biology. From 1989 through 1995, a Head of the Lab of Clin Immunol, Helmholtz Eye Res Inst in Moscow. From 1995 through 2004, a Chair of Dept for Clini Imunol, Moscow Clin Res Inst (MONIKI). Trained at: NIH; Wills Eye Hospital, PA, Univ of Florida in Gainesville; UCSF; J Hopkins Univ, Baltimore, USA. He was an Exe Secretary-in-Chief of the Edit Board, Biomedical Science, a Journal published by the USSR Acad of Sci and the Royal Society of Chemistry, UK. At present, a Chair, Dept for Transl Med, MEPhI, and Director, Center for Personalized Medicine, Sechenov University, Russia. A member of: NY Acad of Sci,; American Chemical Society (ACS); American Heart Association (AHA); EPMA, Brussels, EU; ARVO, ISER, PMC, USA.
Abstract:
A new systems approach to diseased states and wellness result in a new branch in the healthcare services, namely, personalized medicine (PM). To achieve the implementation of PM concept into the daily practice including clinical cardiology, it is necessary to create a fundamentally new strategy based upon the subclinical recognition of bioindicators (biopredictors and biomarkers) of hidden abnormalities long before the disease clinically manifests itself.
Each decision-maker values the impact of their decision to use PM on their own budget and well-being, which may not necessarily be optimal for society as a whole. It would be extremely useful to integrate data harvesting from different databanks for applications such as prediction and personalization of further treatment to thus provide more tailored measures for the patients and persons-at-risk resulting in improved outcomes whilst securing the healthy state and wellness, reduced adverse events, and more cost effective use of health care resources. One of the most advanced areas in cardiology is atherosclerosis, cardiovascular and coronary disorders as well as in myocarditis. A lack of medical guidelines has been identified by the majority of responders as the predominant barrier for adoption, indicating a need for the development of best practices and guidelines to support the implementation of PM into the daily practice of cardiologists!
Implementation of PM requires a lot before the current model “physician-patient” could be gradually displaced by a new model “medical advisor-healthy person-at-risk”. This is the reason for developing global scientific, clinical, social, and educational projects in the area of PM to elicit the content of the new branch.
Kenneth J. Rotondo
President & Founder, Mind Genomics Advisors, USA
Title: Machine Learning & Patient Communication: A Primer for the Non-Data Scientist

Biography:
Ken Rotondo is an authority on consumer experience engineering, client/patient communication, and empirical messaging insights. He has served on the Advisory Counsel to the Dean at the Veterinary College, Cornell University, is a Past President of the New York State Veterinary Medical Society, and represented New York to the AVMA. Currently he is an annual guest lecturer at the Wharton Business School, RPI Lally School of Business, Villanova, Cornell University and Skidmore College. He serves on the Canadian Bio-Alliance Mentoring Board, the AVMA “Futures Commission”, and several not-for-profit boards. As President of Mind Genomics Advisors he has presented at IBM Watson Conference, IBM THINK, Social Media Week NYC, and a variety of medical and professional conferences.
Abstract:
Statement of the Problem: With the growth of personalized medicine, we know that a single drug will not be effective to the same degree in everyone, so why do we still believe that the same singular message would have the same effect for everyone? Day after day, the health care community struggles to effectively deliver critical messages to patients. In many areas of medicine, we can offer precise tailored and effective therapies, but in our attempts to communicate recommendations to patients, we lack the same specificity. Methodology & Theoretical Orientation: Our over-arching goal is to improve health-related communications and messaging resulting in greater adherence, fewer wasted resources and improved outcomes for patients. A patient communication tool, operating at scale, has been developed to identify individual patient “mind-sets” specific to a medical condition. Findings: This presentation will describe a case study in colon cancer screening compliance. The investigation focuses on colon cancer screening because it is commonly recommended, has tremendous life-saving potential and each case of colon cancer is estimated to create a financial burden of nearly $250,000. Conclusion & Significance: The promise of “machine learning” is to gain insights to improve prediction accuracy and increase the probability of a desired outcome. An actionable medical communication methodology will be demonstrated that promotes and encourages healthy behaviors through scientifically-based, patient centered, tailored messaging based on individual mindsets. Applying basic machine learning principles to communication attendees will learn to better understand the variability of patient motivations specific to decision-making and choice.

Biography:
Founder & CEO of R&D Ribitzky, Dr. Ron is a Physician executive with extensive experience in the convergent healthcare IT, life-sciences informatics, and Precision Medicine. He worked for and with global leading brands and world-class organizations, R&D, and academia in the U.S. and 23 developed and emerging economy countries; in addition to ten years in clinical practice. Previously, he co-founded SPH Analytics, and served as a Senior Healthcare Strategist at Intel, VP Advanced Research at Eclipsys, CIO at UMass Medical Center, among others. He published, presented, and led workshops around the world on technology innovation and practice in industry events, public sector, and academia; and held academic appointments at Harvard, UMass, Emory, Weizmann Institute of Science (Israel), Kigali Institute of Science & Technology and Kigali Health Institute (Rwanda). He is a member of HIMSS Blockchain Work Group, and Strategic Advisor to Blockchain start-ups ARNA Genomics, BitMed, and Cryptfunder, among others.
Abstract:
The Total Addressable Market (TAM) for Precision Medicine is on a trajectory of 10%+ CAGR to surpass $100B around 2025 as patient demand is on the rise, the cost of human genome sequencing continues to go down, AI happens deep inside the chip, blockchain is heading everywhere, and medical devices are just about to switch channel to IoT chat. Yet, it still takes 10+ years for basic science discovery to make a difference in the life of the first consumer it was intended for. In this session, we will re-examine the ever-elusive Precision Medicine Paradox, white space R&D, healthcare in disruption, and We the Consumers.
Satish P. RamachandraRao
The University of Trans-Disciplinary Health Sciences and Technology
Title: Exosomes and Precision Delivery of Personalized, Preventive Medicine

Biography:
Rao received his Ph. D. in Neurochemistry from the Christian Medical College, Vellore, India. He has performed postdoctoral studies in Indian Institute of Science, Bangalore, India. He was recruited to Department of Medicine/Nephrology, Thomas Jefferson University, Philadelphia, USA to work on kidney organ function dysfunction biomarkers specifically in subjects with diabetes. In 2007 he moved to University of California at San Diego School of Medicine, California, USA, as an Assistant Professor of Medicine, where he continued his Kidney disease related work in Nephrology. After setting up 2 independently functional laboratories completely from ground up in these two universities in USA, he changed his focus to Clinical Biomarkers of Acute Kidney Injury in the NIH-funded George O’Brien Center for AKI Research at UC San Diego, as the Director of the Biomarkers Laboratory. He moved to Kolar Medical College in 2016 as Professor and Head of the department of Cell Biology & Molecular Genetics, Director of the Center for Exosome Research and the Research Officer of the University, along with multiple translational research collaborations in about a dozen institutions across the country. He continues his association with his previous institution as a Visiting Professor at University of California, San Diego. Recently he has assumed the leadership of a Bangalore-based consortium to develop programs for preventing dengue (ICMR-INFOSYS Initiative for Dengue Vaccine Trial Site Development), and biomarkers of health and wellness in Indian Population at the interface of Western Medicine and Integrative Medicine. His work has been continuously funded by USA-based public health agencies such as National Institutes of Health, Bethesda, Maryland, Juvenile Diabetes Research Foundation, New York. One of his research interests is developing non-invasive technology platforms, such as urine exosome biomarkers of kidney physiology in health and disease.
Abstract:
Multiple clinical and para-clinical research projects carried out at our Biomarkers Laboratory of the O’Brien Center for Acute Kidney Injury Research, UC San Diego School of Medicine demonstrate that the urinary exosome protein content of an individual is rather specific to the health or disease etiology of the subject. For the purpose of this personalized medicine conference, we have focused on one of these etiologies, namely Coronary Artery Bypass Grafting (CABG). Between 4 to 15 days after cardiac surgery insult, 4 out of 10 CABG surgeries result in Acute Kidney Injury (AKI). AKI is determined by elevation in serum creatinine (Scr). Scr elevation can only indicate kidney injury that has already occurred. Therefore, using only Scr elevation is of limited utility in either prediction or prevention of AKI episodes. Presently, AKI prevention in the clinic is thus an unmet clinical necessity.
Our data show that urinary exosome protein content can potentially discriminate between who among the cardiac surgery patients will develop AKI versus who will not, even before Scr elevation has taken place. We reason that this provides the clinician with an opportunity to help the patient prevent AKI phenotype precipitation. In this talk / meeting, the following will be highlighted: (a) how to utilize and develop this clinical opportunity into personalized preventive medicine strategy, (b) the constituent elements of this strategy, (c) the role of personalized omic integration analysis, (d) how to couple it with preventive therapy in principle guided by specific exosome biomarker analysis, and finally (e) the role of big data in precision delivery of personalized and preventive medicine. Further in addition, how exosome biomarker specificity can be expanded and utilized to achieve this precision delivery in other diseases or in healthy individuals in general, will be discussed.

Biography:
Hornshaw is the Director of Scientific Marketing at Metabolon. Before joining Metabolon he worked for Thermo Fisher Scientific and Applied Biosystems-MDS Sciex in roles ranging from a scientist working at the bench through to leading a large team of scientists supporting customers in the field using mass spectrometry. At Metabolon Dr Hornshaw’s goal is to educate the public in general as well as scientists specifically as to the power of mass spectrometry based metabolomics to identify underlying mechanisms in biology affecting health and disease such as how the microbiome communicates with its host, in biomarker discovery and validation, as a tool to enable both precision and personalized medicine and more. Dr Hornshaw has many years of experience as a scientist and of managing scientists working with the ‘mass spec OMICS’ of metabolomics, lipidomics and proteomics as well as clinical and diagnostic applications of mass spectrometry.
Abstract:
We have heard much about genomic medicine but genotype does not equate to phenotype. A potential future of effective precision medicine requires not only estimates of lifetime risk of disease but also accurate measurement of phenotype: molecular phenotype. This will enable assessment of disease risk in the near term, disease progress, effect of intervention on disease progression and so on. Metabolomics enables measurement of molecular phenotype in the form of the assessment of an individual’s overall metabolism (approx 1000 metabolites in plasma or serum) and changes to metabolism, not just individual metabolite measurements, in samples such as plasma, urine and tissue. When utilized in a ‘Systems Medicine’ approach metabolomics becomes a powerful indicator of health and disease. A pragmatic systems medicine approach would be to merge genomics data with large scale phenotypic measurements such as metabolomics, proteomics and imaging. A global metabolomics platform and its use in recent years in population health studies and applying knowledge of metabolism to precision medicine research will be discussed. These areas include diagnosis of inherited metabolic disease, undiagnosed illness, assessment of penetrance, drug toxicity and ‘scientific wellness’.

Biography:
Sergey Suchkov was born in the City of Astrakhan, Russia, in a dynasty of medical doctors, graduated from Astrakhan State Medical University and was awarded with MD. Then maintained his PhD and Doctor’s Degree. And later was working for Helmholtz Eye Research Institute and Moscow Regional Clinical Research Institute (MONIKI). Dr Suchkov was a Secretary-in-Chief of the Editorial Board, Biomedical Science, an international journal published jointly by the USSR Academy of Sciences and the Royal Society of Chemistry, UK. At present, Dr Sergey Suchkov is: (i) a Director, Center for Personalized Medicine, Sechenov University, (ii) Chair, Dept for Translational Medicine, Moscow Engineering Physical University (MAPhI), and (iii) Secretary General, United Cultural Convention (UCC), Cambridge, UK. A Member of the: New York Academy of Sciences, American Chemical Society (ACS), American Heart Association (AHA), AMEE, Dundee, UK; EPMA, Brussels, EU; PMC, Washington, DC, USA and ISPM, Tokyo, Japan.
Abstract:
A new systems approach to diseased states and wellness result in a new branch in the healthcare services, namely, personalized medicine (PM). To achieve the implementation of PM concept into the daily practice including clinical cardiology, it is necessary to create a fundamentally new strategy based upon the subclinical recognition of bioindicators (biopredictors and biomarkers) of hidden abnormalities long before the disease clinically manifests itself.
Each decision-maker values the impact of their decision to use PM on their own budget and well-being, which may not necessarily be optimal for society as a whole. It would be extremely useful to integrate data harvesting from different databanks for applications such as prediction and personalization of further treatment to thus provide more tailored measures for the patients and persons-at-risk resulting in improved outcomes whilst securing the healthy state and wellness, reduced adverse events, and more cost effective use of health care resources. One of the most advanced areas in cardiology is atherosclerosis, cardiovascular and coronary disorders as well as in yocarditis. A lack of medical guidelines has been identified by the majority of responders as the predominant barrier for adoption, indicating a need for the development of best practices and guidelines to support the implementation of PM into the daily practice of cardiologists!
Implementation of PM requires a lot before the current model “physician-patient” could be gradually displaced by a “medical advisor-healthy person-at-risk” model. This is the reason for developing global scientific, clinical, social, and educational projects in the area of PM to elicit the content of the new branch.

Biography:
Ana Sabater studied IT at the University of Barcerlona, Spain in 1997. She has a Maser degree in Marketing from the Universitat of Barcelona. She has been for the past 14 years involved in the IT business related to the health care. She co-founded EUGENOMIC in 2008, where her role has been to develop and put to the market g-Nomic® pharmacogenetics interpretation software to correctly apply pharmacogenetics. She teaches practical applications of pharmacogenetics at the University of Barcelona. Has given more than 30 international keynote presentations about pharmacogenetics and has designed the training program that has been running now for more than 5 years.
Abstract:
A single word Pharmacogenetics is the answer for ‘why certain drugs produce toxic effects to some people & why in certain individuals, a particular drug does not help?’
Knowing the genetic polymorphisms of a patient shows whether a drug will make the intended effect according to the dosage from clinical trials, will require more or less dose, or it should be avoided and seek a therapeutic alternative.In general, patients are polymedicated, however the effect of various drugs administered together may be different (produce ineffectiveness or toxicity), different to what would happen when administered alone. Sometimes, even if a person could take the drugs individually according to the genetics, the drugs themselves could inhibit or induce the other.To successfully apply pharmacogenetics and make a prescription safely and accurately, many parameters should be taken into account, such as drug-drug interactions, drug-lifestyle, inhibitions and inductions and dose variation according to patient studied genes. All this information can only be interpreted together, using a pharmacogenetics interpretation software like g-Nomic®. g-Nomic crosses information concerning prescribed drugs, patient’s genetic variations to give a personalized report with all necessary information: drug interactions, drug with lifestyle, inhibitions, inductions and dose variation according to the patient's genes. Applied pharmacogenetics can indeed avoid many emergency cases, safe lives and money. But to correctly apply pharmacogenetics, an interpretation software is needed since there are too many variables to consider.

Biography:
Brahma D. Sharma, PhD is a retired chemist from R&D and manufacturing of pharmaceutical and medical device industry. He is an author, inventor, scientist, entrepreneur, coach, and speaker. His current book is ‘Cracking the Genetic Code for Prescription Drugs’. His mission to reduce Adverse Drug Reactions (ADR). It’s the fourth leading cause of death in the US after heart attacks, strokes, and cancer. One person dies every 4 minutes. Doctors have been relying on pharmaceutical manufacturers far too long. Drug companies often recommend one dose without considering patients’ genetic variations which impact heavily their drug’s metabolism. One drug/one dose does not fit all. He is educating everyone, patients, doctors, healthcare providers etc. how to reduce ADR by incorporating Pharmacogenetics (PGx) testing in the practice and personalizing prescription drugs that match patient’s hepatic gene variations. ADR is estimated to cost over $136 Billion in the US not counting priceless suffering of patients and their loved ones. He believes that the addition of opioids often begins by the drugs prescribed by a doctor and taken as directed. His goal is for doctors to start with “Let’s start with swabbing your cheek. This will tell me which drugs will do good for you and which drugs may harm you.”
Abstract:
Time is up for continuing to prescribe drugs by trial and error. Eliminate the guess work. Most drugs work when prescribed to a ‘right’ patient.On the other hand, a ‘right’ drug prescribed to a ‘wrong’ patient could be and often is deadly.
The dilemma is: how do you differentiate between ‘right’ and ‘wrong’ patient before prescribing a drug? Even siblings often metabolize drugs differently due to inherited mutations (variations) among genes in their livers. Pharmacogenetics (PGx) is here to guide you. PGx test involves a simple cheek swab, yet provides powerful clinically actionable data for the life of a patient. It predicts before a drug is prescribed, if it is a ‘right’ drug for the ‘right’ patient or it has a high risk of potentially creating Adverse Drug Reaction (ADR) due to a slow acting gene, representing a ‘wrong’ patient, and should be replaced by another therapeutically equivalent drug which is metabolized by another gene which is functioning normally in the patient’s liver. Unmetabolized drug starts accumulating in the body and can lead to ADR due to increased toxicity and side effects, in millions of patients in the US alone. In fact, ADR is now the 4th leading cause of death here, one patient dies every 4 minutes. Insurance companies including Medicare often pay for the test.
Who needs PGx test urgently
It’s too late to order the test during surgeries in ERs. You need to have patient’s PGx report through EMR, EHR or cloud to prescribe personalized medicine to control pain, bleeding etc. I recommend every newborn should have the PGx test to insure even first prescription the child will ever get be personalized to reduce potential of an ADR. The risk of ADR increases exponentially by the number of prescription the patient is on. Coumadin/Warfarin, a commonly prescribed blood thinner to control thrombosis is number one drug requiring ER visits among senior patients.
PGx reducing healthcare cost
University of Illinois Health’s Personalized Medicine Program saved $600K annually by reducing hospital readmissions when they used PGx test to guide dosing of only one drug, Coumadin/Warfarin.
FDA requiring PGx test
FDA along with EMA, PDMA and HCSC is recommending, in fact requiring a PGx test before prescribing 54 of the drugs carrying Black Box Warnings. Alas, healthcare professionals keep ignoring the requirement. I appeal to all healthcare professionals to be progressive and adopt PGx test and change their practices to personalized medicine.

Biography:
Gil Blander is internationally recognized for his research in the basic biology of aging and translating research discoveries into new ways of detecting and preventing age-related conditions. He leads a team of biology, nutrition & exercise physiology experts, and computer scientists at InsideTracker. He received a Ph.D. in biology from the Weizmann Institute of Science and completed his Post Doctoral fellowship at MIT, before going on to found InsideTracker. The InsideTracker platform analyzes key biochemical and physiological markers and applies algorithms and large scientific databases to determine optimal zones for each marker. The system then provides nutrition, exercise, supplements and lifestyle interventions that empower people to optimize their markers, increasing vitality, improving overall health, as well as athletic performance and extending life.
Abstract:
The trend toward personalized approaches to health and medicine has resulted in a need to collect high-dimensional datasets on individuals from a wide variety of populations, in order to generate customized intervention strategies. However, it is not always clear whether insights derived from studies in patient populations or in controlled trial settings are transferable to individuals in the general population. To address this issue, an observational analysis was conducted on blood biomarker data from 1033 generally healthy individuals who used an automated, web-based personalized nutrition and lifestyle platform. Using the resulting dataset, a correlation network was constructed to generate biological hypotheses that are relevant to researchers and, potentially, to users of personalized wellness tools. The correlation network revealed expected patterns, such as the established relationships between blood lipid levels, and novel insights, such as a connection between neutrophil and triglyceride concentrations that has been suggested as a relevant indicator of cardiovascular risk. Biomarker changes were assessed from baseline to follow-up, relative to platform use. Preliminary associations were found between the selection of specific nutrition and lifestyle interventions and biomarker outcomes. Across many biomarkers measured, there was a significant trend toward normalcy in participants whose biomarker values were out-of-range at baseline.

Biography:
Simon came to Biologics Consulting with over eleven years of experience at the FDA. During her time at FDA she served as a Master Scientific Reviewer in the Office of In Vitro Diagnostics and Radiological Health (OIR). Dr. Simon has been a lead reviewer of many pre-market applications for IVD devices including PMA, 510(k), Pre-IDE, IDE and Pre-submissions. She has served as an FDA speaker in public forums on a diverse array of topics, including the de novo process, pharmacogenomics and personalized medicine, and premarket IVD submissions. Dr. Simon holds a Ph.D. in Microbiology from the University of Virginia.
Abstract:
This presentation by a former FDA reviewer will provide an overview of FDA regulation of in vitro diagnostic devices (IVDs). This presentation will help audience members make the leap from discovering a new biomarker to marketing an FDA approved or cleared diagnostic test. Design of appropriate analytical and clinical validation studies to support subsequent IVD data submissions to FDA will be covered, as well as best practices for preparing an FDA premarket submission. Seasoned professionals as well as those new to in vitro diagnostics will benefit from hearing about IVD device regulation from a former FDA reviewer. This will better enable them to prepare for IVD device clinical trials and subsequent data submission to FDA.

Biography:
Jain is a neurologist/neurosurgeon with career in North America. He holds fellowships in neurosurgery from the Royal Colleges of Surgeons in Canada and Australia. After retirement from neurosurgery, he started a second career in pharmaceutical medicine and biotechnology in Switzerland. Currently he is a Fellow of the Faculty of Pharmaceutical Medicine of the Royal College of Physicians of UK. In addition to work as a consultant in neurology, he is also involved in biotechnology. His company, Jain PharmaBiotech is involved in Research and publications on biotechnology and applications in pharmaceuticals and healthcare. He is integrating advances in biotechnology and other areas to advance personalized medicine. He has given several invited lectures and conducted workshops on personalized medicine since 2000 in Asia, Europe, and USA. He served as a visiting professor personalized medicine at the University of Kazakhstan at Almaty in April 2018. Jain is developing personalized medicine since 1998, and wrote the first monograph on this topic, which evolved into “Textbook of Personalized Medicine”, 2nd edition (Springer 2015). His 465 publications, besides several papers on personalized medicine, include 30 books (6 as editor and 25 as the author). Important authored books include “Applications of Biotechnology in Neurology” (Springer, 2013), “Applications of Biotechnology in Oncology” (Springer, 2014) “Handbook of Biomarkers”, 2nd ed (Springer 2017), and “Handbook of Neuroprotection”, 2n ed (Springer 2018, in preparation). He has edited “Applied Neurogenomics” (Springer 2015). Editorial board memberships of journals include “Technology in Cancer Research & Treatment and “Nanomedicine” (London).
Abstract:
Personalized neurology is the application of principles of personalized medicine, i.e., the prescription of specific therapeutics best suited for an individual taking into consideration both genetic, epigenetic, and environmental factors that influence response to therapy. The aim is to improve efficacy and reduce adverse effects of therapy. Personalization of therapies for neurological disorders is based on a better understanding of the disease at the molecular level. Molecular diagnostics, molecular neuroimaging, sequencing and monitoring of gene expression by microarrays are important technologies for this purpose. Besides omics, e.g., neurogenomics and neuroproteomics, nongenomic technologies such as nanobiotechnology are also used. Biological therapies for neurological disorders, such as cell therapy, gene therapy, gene editing, RNAi, vaccines and monoclonal antibodies can also be personalized. Biomarkers and integration of diagnostics with therapeutics are important for the selection and monitoring of treatments. Biomarkers enable presymptomatic diagnosis, selection of appropriate treatment, assessment of disease progression, and evaluation of patient response to therapy. Nanobiotechnology has refined molecular diagnosis, improved drug formulation and targeted delivery through the blood-brain barrier to the lesion in the brain and spare the normal tissues to reduce systemic toxicity. Integration of multiple factors into personalized approach requires use of bioinformatics. A personalized approach can be incorporated in algorithms for the management of various neurologic disorders. Examples of neurological disorders will include Alzheimer disease, Parkinson disease, epilepsy, and stroke. Advantages and future of personalized neurology are:
- The availability of low-cost genomic sequencing will expand the use of genomic information in the practice of neurology.
- Sequencing of the genome is enabling genetic redefinition of several neurologic disorders as well as better insight into their pathomechanisms to improve our understanding and facilitate early detection by molecular methods.
- An increase in the ability to anticipate diseases rather than just reacting to them after onset may enable the institution of preventive measures.
- The precision and effectiveness of drugs is increased.
- Drugs can be better targeted to diseases in some patients based on genotype information.
- Development of more effective personalized medicines may obviate the need for surgery in some chronic neurological disorders.
Ghulam Rasool Mashori
Peoples University of Medical & Health Sciences for Women
Title: Effects of Nifedipine An Antihypertensive Drug on Glucose and Epinephrine.

Biography:
Ghulam Rasool Mashori is working as Professor, Peoples University of Medical & Health Sciences for Women, Nawabshah, Pakistan supervises to Graduate & Postgraduates fellows. Dr Mashori after graduation acquired PhD (Pharmacology) in 1994 from the Faculty of Medicine, University Kebangsaan Malaysia. He completed many Short/Long Courses and Trainings in Research areas of Health Sciences & Management Sciences. Mr. Mashori has been involved in Teaching/ Trainings to MBBS & PharmD students and Research/ Supervisory works at MPhil & PhD levels with various Institutions i.e. Jinnah Postgraduate Medical Center, Karachi, Pakistan etc., produced many research fellows. Beside this he has experiences in Research, Quality Control & Administration with NIH, Islamabad, as Deputy Director General (Pharmacovigilance) and as Director, Central Drugs Laboratory with Ministry of Health, Government of Pakistan. He has Supervisory experiences with reputed Pharma Industries. He holds Research, Managerial, and Administrative skills. 38 research papers are on his credit published in reputed Journals. He is also Editorial/Review Board Member of various world’s reputed Journals. Dr Mashori delivered many talks on International Forums in various conferences/ workshops held in Pakistan, Malaysia, Hong Kong, Singapore, USA & Europe. His field of interest is Cardiovascular Pharmacology, Diabetes, Endocrinology, Neuropharmacology, Genetics etc.
Abstract:
Hypertension, hyperlipidemia, Hyperglycemia and Cigarette smoking are major risk factors for the development of Coronary Heart Diseases (Newcomer & Hennekens, 2007). Hypertension is the leading curable risk factor for cardiovascular disease, affecting more than 1 billion people worldwide (Patel et. al 2016). Treatment of hypertension is to reduce future cardiovascular morbidity and early death. Antihypertensive therapy has shown improvement in the incidences of stroke, heart failure & renal failure however, the incidence of CHD has not been reduced to that degree (The Sprint Research Group, 2017). Normal human BP (Systolic/Diastolic) ranges from 120/80mmHg to 140/90mmHg. Hypertension is the continuous elevation of resting systolic ≥140mmHg & diastolic ≥90mmHg BP (Carretero & Operil, 2000). Effects of Nifedipine an antihypertensive drug were seen on Glucose, Insulin & Epinephrine, in the light of Helsinki Criteria, a study designed on nondiabetic and diabetic hypertensive patients. Two sets, minimum of 12 patients (6-non-diabetics and 6-diabetics) were recruited for treatment. In Set-A non-diabetic hypertensive patients were kept on placebo first for two weeks and GTT was performed, then patients switched on Nifedipine at an average dose 23.8±7.4mg/day. When the diastolic BP normalized ≤90mmHg at-least for 6-weeks period, then second GTT was performed. In Set-B, diabetic hypertensive patients treated with Nifedipine first at an average dose 21.3±3.5mg/day for 6-weeks period, GTT was conducted, followed by a placebo period for 2-weeks then the patients went through second GTT for quantifying glucose, insulin and catecholamine blood levels and results were compared. Based on results it is concluded that, Nifedipine worsens Glucose Tolerance and it is a diabetogenic drug.
Larina Vera
Russian national research medical university
Title: Problems and perspectives of personalized medicine

Biography:
Larina Vera, PhD, MD, professor of the department of outpatient medicine of Russian national research medical university named after N. I. Pirogov. She has published more than 100 papers in Russian journals and is a member of the editorial board.
Abstract:
In Russia, as in many countries around the world, there was an individualized and paternalistic approach to the treatment of the patient. In the past, it was based on the personal doctor’s experience, the doctor-patient relationship and often the patient's socioeconomic status. The priority of individualized treatment began to give way to clinical guidelines based on evidence-based medicine in relation to the introduction of the principles of "industrialized" and evidence-based medicine into practice. At the same time, over the past decade it has become clear that such approaches cannot resolve the complex of problems associated with the patient's "burden of disease".
Awareness of the need to minimize the burden of treatment for the patient, his family and the health care system has emerged in relation to the development of new technologies, increased longevity, increase patients number with multimorbidity and fragility, emergence of research results indicating that the same method of treatment (drug) can affect the patient in different ways. More and more specialists are talking about the need for individualized approach in the choice of tactics for managing the patient, taking into account the biological characteristics, the possibilities of medicine and many other non-medical factors (ethical, social, financial).
A lack of knowledge is a significant problem for the introduction into the clinical practice of personalized medicine despite of their geometric growth at the present time. In fact, there are no randomized studies, that all the way of treatment and diagnostic methods with an infinite variability of multimorbidity are evaluated, there is no possibility to use in clinical work all the accumulated knowledge in the world (this the limited capabilities of a doctor and even a multidisciplinary team; lack of a common language of communication, a database of such information, rapid access to it, taking into account the individual needs of the patient), only the beginning of the selection of treatment taking into account genetic and epigenetic factors.
Tori Strong
Vyripharm Biopharmaceuticals,USA
Title: Novel Formulation of Cannabinoid Analogues Treating DLBCL and MCL

Biography:
Tori Strong has completed his PhD from the University of Texas Medical Branch at Galveston. He is the director of Intellectual Property and Technology at Vyripharm Biopharmaceuticals, a premier biotech organization. He has served as a Patent Examiner for the USPTO and is now directly involved in the technology development to commercialization which includes strategies of building intellectual propery.
Abstract:
Novel Formulation of Cannabinoid Analogues Treating DLBCL and MCL: Diffuse large B-cell lymphoma (DLBCL) and mantle cell lymphoma (MCL) represent the most common and most aggressive forms of Non-Hodgkin lymphoma (NHL) respectively. With CB1 antagonists as potential therapeutics for both DLBCL and MCL, we formulated VYR-206, developed from existing obesity treatment Rimonabant by the addition of our tetraazacyclic (N4) conjugate derivative. This allows the potential for image guided theranostic application for diagnosis, precision and assessment of therapeutic response through radiotracer chelation. Our study is aimed at demonstrating VYR-206 activity in DLBCL and MCL for sensitivity or resistance. Cells from representative DLBCL and MCL cell lines were plated at 5,000 cells per well. The cells were incubated for 72 hours in 20 µL medium with 10% FBS and varied concentrations of experimental cannabinoid antagonist VYR-206, Rimonabant, or dimethylsulfoxide (DMSO). Viability assays were conducted using Celltiter-Glo Luminescent Cell Viability Assay. Experiments were performed 2-3 times independently, with concentration tested in triplicate. Most DLBCL cell lines treated with VYR-206 had a reduction in viability at concentrations of 50μM or greater with few cells line displaying limited response even at concentrations of 100μM. Increased variability is seen among MCL cell lines treated with VYR-206, most having a reduction of viability at concentrations of 25μM or greater, with few cell lines at concentrations of 50μM and 2 cell lines showing no response even at concentrations of 100μM. The discrepancy in response in both DLBCL and MCL may be due to genetic variability among cell lines.
Romina Ortiz
Rare Genomics Institute,USA
Title: Lifting the burden of Rare Disease by shortening the Diagnostic Oddyssey

Biography:
Abstract:
From the Orphan Drug Act of 1983, a rare disease is a condition that affects fewer than 200,000 people in the United States (1). In the European Union, the condition must only affect fewer than 1 in 2,000 people (2). While the numbers seem small, there is an estimated 350 million people that suffer from rare diseases, with 25-30 million belonging to the US alone and so far there have been over 7,000 different rare diseases identified (1). To put this in perspective, there are more Americans affected by rare disease than for HIV, Heart Disease or Stroke combined (3). It is important to understand that by nature rare diseases are difficult to diagnose, and consequently are not tracked. Thus, it is hard to accurately determine the number of rare diseases and their impact on a population. The average length of time from onset of symptoms to an accurate rare disease diagnosis is nearly 5 years, and patients see an average of over 7 different physicians before a diagnosis is made (5).This delay in diagnosis results in chronic physical, emotional and socioeconomic burden to both the patient and their family. A European Cost of Illness Study interrogating published literature on the cost of 10 selected rare diseases found that overall, the availability of data on economic burden for rare diseases was correlated with the availability of therapies, not the severity of the disease. Also, most rare diseases reviewed were found to have significant economic burden and indirect costs ( many associated with loss of productivity) exceeded the level of direct costs (5). Rare Genomics has served over 500 undiagnosed patients since 2011, helping them access next generation sequencing to accelerate their pathway to a cure. We have seen the same patterns reported for rare diseases in our own patients including heterogeneous disease marked by a range of severity across a variety of biological systems. The most common systems affected are Neurologic, Respiratory, Gastrointestinal, Muscular and Cardiovascular. The average RG patient has also seen a range of physicians, the top three specialties consulted are: Neurologist, Clinical Geneticist, Opthamologist and Gastroenterologist.Lastly, undiagnosed/rare disease patients typically have already undergone a gamete of testing, the most common tests are: MRI, DNA Microarray and Single/Panel Sequencing. Because 80% of rare disease are genetic in origin, we hope that by providing support and access to next generation sequencing, we can help reduce the time and burden these families must undergo before identifying appropriate treatment for their disease.

Biography:
Andreas Weiler is the Global Business Unit Head of Emerging Technologies at Lonza where he oversees the Cell and Gene Therapy Contract Manufacturing businesses. Before joining Lonza he worked for Sigma Aldrich (SAFC), another leading CDMO (Contract Development and Manufacturing Organization) to the Pharma/biotech industries, for over 17 years where he held several positions as Business Director and finally as the Head of Strategic Marketing to the organization. Since 2015, when Andreas joined Lonza, Cell and Gene Therapy sales have more than doubled and the manufacturing footprint expanded significantly to cover the globe with facilities in North America, Europe and Asia. Andreas holds a PhD in Organic Chemistry from University of Freiburg and an MBA degree.
Abstract:
Cell and Gene Therapies are seen as the next frontier in medicine as they have the potential to bring cures to patients that suffer from life-threatening diseases without many treatment options available to them. The idea that a patient’s own cells can be reprogrammed to replace or eliminate faulty genes or to attack cancer cells in a way that is not naturally possible is giving hope to many. In 2017, we have seen several approvals from these innovative medicines, Kymriah, Yescarta and Luxturna to name a few. However, a key challenge for all players and drug-makers in this field remains to be addressed: the cost of manufacturing is critically high due to the nature of these highly personalized medicines. This high cost greatly limits the number of patients that can be eligible and threatens the sustainability of the therapy as a whole, and this is likely to be reflected in the already high price tags of these drugs for the foreseeable future. In his presentation, Andreas Weiler would like to show the way these medicines are manufactured and delivered to the patients could have significant and disruptive implications for the future of healthcare. He will be sharing his vision and innovative technologies that could reduce the cost of these medicines significantly and make them accessible to a larger number of patients.

Biography:
Jacques Beckmann is the Head of clinical bioinformatics at the Swiss Institute of Bioinformatics (SIB) from December 2012 till December 2016. He served previously for ten years as Professor and Chairman of the Department of Medical Genetics at the Faculty of Biology and Medicine of the Univ. of Lausanne as well as head of the Medical Genetics Service of the Centre Hospitalier Universitaire Vaudois (CHUV). Previously, he held a chair as Professor at the Dept of Molecular Genetics at the Weizmann Inst of Science, Rehovot, Israel. Initially trained in molecular genetics he then moved to genetics. In the 1980s, together with Prof. M. Soller from the Hebrew Univ, they pioneered the use of marker-assisted genetic improvement in plants and animals, focusing on Quantitative Trait Loci (QTLs). His interest shifted in 1990 to human genetics with a move to Paris, where he held successively senior research positions at the CEPH, Généthon (Evry), and finally the Centre National de Génotypage (CNG, Evry), where he was Deputy-Director. During those years he collaborated with Prof. D. Cohen, J. Weissenbach, M. Lathrop, J. Dausset and others and contributed significantly to the elaboration of genetic, physical and gene maps of the human genome, as well as to the positional cloning of a number of disease loci, many of which are involved in muscular dystrophy. Prof. Beckmann has published over 390 scientific peer-reviewed articles in molecular genetics, genetics and genomics, and has an ISI H-index of 86. He has served on the editorial boards of a number of scientific journals. His research interests include genomic disorders, pharmacogenetics as well as the genetic basis of complex traits.
Abstract:
The recent years have seen the emergence of both ground-breaking scientific developments in high-resolution, high-throughput data gathering technologies enabling cost-effective collection and analysis of huge, disparate datasets on individual health, as well as of sophisticated clinical bioinformatics or machine learning tools required for the analyses and interpretation of this wealth of data. These developments have triggered numerous initiatives in precision medicine (PM), a data-driven and currently still, essentially a highly genome-centric initiative. Proper and effective delivery of PM poses numerous challenges. Foremost, PM needs to be contrasted with the powerful and widely used practice of evidence-based medicine (EBM). The latter is informed by meta-analyses or group-centered studies from which mean recommendations are derived. These amount at first approximation to a “one size fits all” approach, whose major limit is that it does not provide adequate solutions for outliers. Yet, we are all outliers for one or another trait. In contrast to EBM, one of the strengths of PM, which focuses on the individual, lies in the area of individualized management, and this includes outliers.To achieve these objectives, it will be necessary to bridge PM and EBM. Through the collection, analyses and sharing of standardized medically relevant data globally, evidence-based PM will shift progressively from therapy to prevention, thus leading eventually to improved, clinician-to-patient communication, citizen-centered healthcare and sustained well-being. We will discuss challenges and opportunities towards these goals.
Stephen Harlin
The Harlin Center for Genomic Medicine, USA
Title: Implementing Genomics in a Precision Medicine Practice

Biography:
Abstract:
A tremendous opportunity exists for implementing genomics into clinical practice. However, there is little research on genomics-based practice protocols or clinical decision support systems (CDSS’s) that incorporate genomic information technologies at the point-of-care.
We developed a CDSS whose user interface combines raw DNA sequence variant data and gut microbial biomarkers to enable a high level, holistic interpretation of the holobiome. Our goal was to individualize therapeutic and preventive interventions, based on insights gleaned from two ‘omics’ data sources and advanced laboratory biomarkers for oxidative stress and inflammation.
While several studies have shown that polygenic risk analysis accurately predicts the individual risk of developing a chronic disease, our model attempts to extend risk stratification to identify molecularly defined subsets of individuals. Specifically, our CDSS clusters polymorphisms in multiple genetic loci that contribute to the pathogenesis of many diseases: glucose and lipid dysregulation, endothelial dysfunction, mitochondrial stress, deficiencies in DNA repair capacity, circadian disruption, maladaptive emotion regulation, and a dysbiotic gut microbiome.
We believe that intelligent decision support tools are crucial to the integration of heterogenous inputs from ‘omic’ technologies and evidence-based clinical regimens. Towards that end, we have designed an application that allows for easy, reliable, and rapid assessment of genotype-phenotype relationships. Output is relevant and intelligible to both physicians and patients. In short, our vision of a genomic CDSS fully embraces the concept of the clinical holobiome.
Finally, we present a descriptive case study of a 58-year-old woman with a history of thrice recurrent bladder cancer. The presentation demonstrates how genetic loci, likely representing disease mechanistic pathways, can be paired with microbiota datasets and interpreted in the context of other known risk factors. This comprehensive approach provides new prospects for disease management and prevention.
Rizwan Ya. Abdullaiev
Kharkiv National Medical University, Ukraine
Title: Transvaginal ultrasound characteristics of the uterus and ovaries in young women with polycystic ovaries

Biography:
Rizvan Abdullaev is currently working in the Kharkiv National Medical University Ukraine. He has published numerous research papers and articles in reputed journals and has various other achievements in the related studies. He has extended his valuable service towards the scientific community with his extensive research work.
Abstract:
The aim of the study was the echographic semiotics of PCOS in young women in a transvaginal triplex mode.
Materials and Methods: The study included the results of transvaginal ultrasound examination 193 female with menstrual irregularities, aged 18–27 years were recruited in the study. Transvaginal echography (TVE) was performed on the 4th-6th and 8th-10th days of the menstrual cycle, and in the absence of menstruation on any day. All women had the ultrasound criteria of PCO: the presence of 12 or more 2-9 mm ovarian follicles and ovarian volume of more than 10cm3. Considering the distribution of follicles in the ovaries and their volume, the blood flow in the stroma, the level of antimulylerovogo hormone, the ratio luteinizing and follicle-stimulating hormones (LH / FSH), polycystic ovary syndrome (PCOS) was diagnosed in 139 (72.0%) women, multifollicular ovaries (MFO) in 54 (28.0%) women.
Results: Peripheral location of ovaries in women with PCOS was noted in 78 (56.1 ± 4.2%) cases, mixed in 61 (43.9 ± 4.2%) cases (P <0.05), in patients with MFO in 23 (42,6 ± 6,7)%) and in 31 (57,4 ± 6,7%) cases, and among the fertility healthy women in 27 (79,4 ± 6,9%) and in 7 (20,6 ± 6.9%) cases respectively (P <0.001). An enhanced in stromal echogenicity was noted in 74 (53.2 ± 4.2%) of women with PCOS, in 17 (31.5 ± 6.3%) women with MFO (P <0.01).
The number of follicles within the limits of 13-15 was registered in 64 (46.0±4.2%) of women with PCOS, 38 (70.4±6.2%) of women with MFO (P<0.001), and more than 15 follicles - in 75 (54.0±4.2%) and in 15 (27.8±6.1%) women (P<0.001), respectively.
In the group of PCOS, the average volume of the ovaries was 18.1±3.8cm3, in the group of MFO – 12.3±2.8cm3, respectively. There were no statistically significant differences in the volumes between these groups. However, in the both groups the volume of the ovaries was significantly (P <0.01) higher than the parameters of the control group.
The vascularization of the ovary stroma in the patients with PCOS was significantly more than in the MFO. The average value of Vmax in the group of PCOS was 49.1±8.6cm/s, in the group of MFO - 35.6±7.1cm/s, in fertility healthy women - 18.9±4.6cm/s, respectively (P<0.001 and P<0.05). Parameters of resistances index (RI) in these groups were 0.48±0.03; 0.54±0.03 and 0.54±0.03, respectively (P<0.05).
The average length of the cervix in the group of women with PCOS was 43.7 ± 3.6 mm, in the group with MFJA - 34.1 ± 3.2 mm, in healthy women - 28.6 ± 2.9 mm, respectively.
The average uterine body length in the group of women with PCOS was 43.7 ± 3.6 mm, in the group with MFJA - 34.1 ± 3.2 mm, in healthy women - 28.6 ± 2.9 mm, respectively.
The ratio of the length of the cervix and the body of the uterus was 1.12 ± 0.07; 0.92 ± 0.06 and - 0.69 ± 0.05, respectively.
Conclusion:
1. Mixed type distribution of follicles, their number is more than 15 on the echographic section, the volume of the ovary is more than 14 cm3, an increase in the number of color vascular signals, an increase in the maximum systolic velocity of more than 50 cm/s, a decrease in peripheral resistance to blood flow of less than 0.51 increases the reliability of ultrasound PCOS criteria.
2. In the syndrome of polycystic ovaries, uterine hypoplasia is observed, which is manifested by an increase in the length of the cervix and an increase in the ratio of the length of the cervix and the body of the uterus.
Getu Molla Tarekegn
Nutrition at Micronutrient Initiative,Ethiopia
Title: Integrating micronutrient interventions into health systems for program sustainability; a case study on vitamin A supplementation (VAS) in Ethiopia.

Biography:
Getu Tarekegn has expertise on research, emergency preparedness & response plan, project design, implementation, monitoring and evaluation, and proposal writing. In line with MI’s strategic objectives in Africa, he contributed to improved child health and nutrition in Ethiopia by supporting the Federal Ministry of Health in the implementation of effective, integrated and sustainable vitamin A supplementation. He has contributed a lot in child survival interventions, community based nutrition program and in the development of nutrition related policies and strategies in Ethiopia. Getu has been coordinating the integration of vitamin A supplementation program from more vertical program into the routine health system in Ethiopia. Considerable advances have been made in integrating VAS along wide range of health system dimensions including policy, financing, planning, training, supply management, supportive supervision and M&E. Children aged 6-59 months that received two doses of vitamin A through integrated VAS delivery strategies has been maintained at above 80%.
Abstract:
Statement of the Problem: Integration of micronutrient interventions into a country’s health system is a means for achieving sustainability of high impact and cost-effective interventions. The biggest challenge with integration is to maintain high coverage achieved through less integrated interventions. To address this concern, Micronutrient Initiative (MI) developed a framework for integrating micronutrient interventions into health systems. The framework was used in the design of a 4-year program (2011-2015) on integrating vitamin A supplementation into the Ethiopian health system to maintain the high VAS coverage which had been achieved through non integrated intervention. The purpose of this paper is to examine the extent to which VAS has been integrated into the Ethiopia health system, & trends in VAS coverage achieved through integrated delivery, and the lessons learnt. Methodology: A situation assessment on the extent of integration of VAS into the Ethiopian health system was done using the MI’s framework for integration to determine the baseline and set targets. The framework was adopted from WHO Health System Building Blocks which includes; service delivery, human resource, procurement and supplies, M&E, financing, Stewardship & Governance. Findings: Strong health system support at all levels allowed relatively high VAS coverage to be maintained while shifting to an integrated routine system. The integration score has improved from 17 to 25 point score during the four years of program implementation. Conclusion & Significance: The MI’s framework for integrating micronutrients into health system has so far successfully guided Ethiopia to design a project that has helped the country to systematically transition from less to more VAS integrated intervention. Government ownership & the clear framework for integration has contributed to the good results achieved so far. Recommendations are made to do a cost analysis to determine the direct costs of reaching children with VAS through integrated intervention.

Biography:
Professional experience in oncology circles, supported by multiple collaborations with major hospitals, allowed LOUIS CAZE to benefit from a reference fame as an Expert Support Care. First, Specialist lymphatic, muscular and circulatory LOUIS CAZE makes the total body approach his chosen field. It captures the benefits of physical-mental balance in the treatment of chronic diseases, and specializes in supportive care dedicated to cancer diseases. His collaboration with renowned organizations such as the Institute Gustave Roussy, was punctuated with exceptional professional and human experience. LOUIS CAZE is involved in implementing a comprehensive approach to cancer patient, in a real project of care that optimizes patient's quality of life, ensuring the different key aspects of health: the well physical and psychological, but also the entire social and family interactions. After these experiences, LOUIS CAZE work for the development of Support Care, including the management of pain and psychology.
Abstract:
What is supportive care?
Support care means all care and support that can be offered to a person suffering from cancer, alongside specific treatments to cure his disease such as chemotherapy, radiotherapy and surgery. They aim to Reduce the impact of disease and treatment. For this, a team of professionals specialized in very different fields put their skills available to patients to help them cope with this difficult time.The support can be offered care during and after treatment of the disease but also when the cancer treatments have no effect. They adapt to the needs of patients and their families.
What needs do they meet? The disease affects all aspects of daily life. The needs that may occur are numerous.
Support care can meet some of these needs:
• Primarily to control the symptoms related to the disease or its treatment
• In case of physical or psychological suffering
• To break isolation
• To learn to live with the physical consequences imposed sometimes disease
• To resume normal course of his life and benefit from the best possible living conditions, and that whatever the chances of recovery
• For many other reasons, each patient with the needs of its own.
Tamer M A Mohamed
Tenaya Therapeutics, USA
Title: Regulation of Cell Cycle to Stimulate Adult Cardiomyocyte Proliferation and Cardiac Regeneration

Biography:
Tamer Mohamed has completed his Ph.D. at the age of 30 years from University of Manchester, UK and held various Postdoctoral training at the University of Manchester, University of Glasgow, and University of Gottingen. He is currently conducting a collaborative research program between the University of Manchester and the J. David Gladstone Research Institute (UCSF) to identify novel therapies for heart failure. He has published more than 15 papers in reputed journals in the past 10 years and has been awarded several awards including the prestigious Young Investigator Award at the European Society of Cardiology Congress in 2010.
Abstract:
Background: Heart failure is often caused by loss of cardiac cells that are unable to re-enter the cell cycle for regeneration. Numerous attempts to identify such cell cycle regulators that could induce cell division of cardiomyocytes, or other cell types, have resulted in nuclear division (karyokinesis), but inefficient cleavage into two distinct daughter cells (cytokinesis) and subsequent survival. Such strategies stimulate cell cycle markers in no more than 1% of cardiomyocytes, limiting their utility.
Methods and results: Here, we took a combinatorial approach to screen for cell cycle factors and conditions that could recapitulate the fetal state of cardiomyocyte division. We found that ectopic introduction of the Cdk1/CyclinB1 and the Cdk4/CyclinD1 complexes promoted cell division in at least 15% of mouse and human cardiomyocytes in vitro. Rigorous assessment of cell division in vivo with the cardiac-specific (a-MHC) Cre-recombinase dependent Mosaic Analysis with Double Markers (MADM) lineage tracing system revealed similar efficiency in adult mouse hearts, leading to cardiac regeneration upon delivery of cell cycle regulators immediately after myocardial infarction and even one week after injury. This ability of cardiac regeneration resulted in significant improvement in cardiac function following acute or subacute myocardial infarction. Intra-myocardial injection of adenoviruses encoding the 4 cell cycle gene either injected at the time of the infarction or one week following myocardial infarction resulted in significant improvement in cardiac function as assessed by Echocardiography and MRI compared to animals received control virus. Furthermore, chemical inhibition of Tgfb and Wee1 made CDK1 and cyclin B dispensable, simplifying the minimal genetic requirement.
Conclusion: These findings reveal a discrete combination of genes that can unlock the proliferative potential in cells that had permanently exited the cell cycle.

Biography:
Ahmed Nasr Ghanem was qualified from Mansoura University, Egypt 1974. Passed is FRCS in 1983 and obtained his MD degree in 1988. He worked as Consultant Urologist Surgeon in Egypt, Saudi Arabia and United Kingdom. He reported over 40 publications. Now he is happily retired in Egypt.
Abstract:
The transurethral prostatectomy syndrome (TURS) is defined as severe vascular hypotension reaction that complicates endoscopic surgery as a result of massive irrigating fluid absorption causing severe acute dilution hyponatraemia (HN) of <120 mmol/l. The vascular shock is usually mistaken for one of the recognized shocks and Volumetric Overload Shock type 1 (VOS1) is overlooked making Volumetric Overload Shock Type 2 (VOS2) unrecognizable. In adults VOS1 is induced by the infusion of 3.5-5 litres of sodium-free fluids and is known as TURS or HN shock. VOS2 is induced by 12-14 litres of sodium-based fluids and is known as the adult respiratory distress syndrome. The most effective treatment for VOS1 and VOS2 is hypertonic sodium of 5%NaCl or 8.4% Sodium Bicarbonate. The literature on TURS is reviewed and the underlying patho-etiology is discussed. As Starling’s law for the capillary-interstitial fluid transfer, which underlies the principles of fluid therapy, proved wrong an alternative mechanism was found by studying the hydrodynamics of the porous orifice (G) tube akin to capillary. Incorporating the G tube in a chamber (C), representing the interstitial space surrounding a capillary, demonstrated a rapid dynamic magnetic field-like fluid circulation between the C and G tube lumen. The G-C phenomenon is autonomous having both filtration and absorption forces making a true replacement for Starling’s law in every tissue and organ of the body.
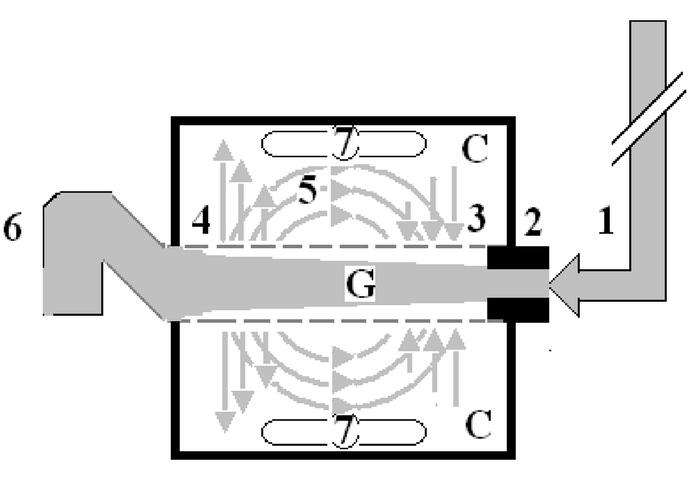
Figure: shows Diagram of the porous orifice (G) tube enclosed in chamber (C) based on several photographs demonstrating the magnetic field-like G-C circulation phenomenon. The proximal inflow (arterial) pressure (1) pushes fluid through the orifice (2) creating fluid jet in the lumen of the G tube. The fluid jet creates negative side pressure gradient causing suction maximal over the proximal half of the G tube near the inlet (3) that sucks fluid into lumen. The side pressure gradient turns positive pushing fluid out of lumen over the distal half maximally near the outlet (4). Thus the fluid around G tube inside C moves in magnetic field-like fluid circulation (5) taking an opposite direction to lumen flow of G. tube. The inflow (arterial) pressure (1) and orifice (2) induce the negative side pressure energy creating the dynamic G-C circulation phenomenon that is rapid, autonomous and efficient in moving fluid out from the G tube lumen at (4), irrigating C at (5), then sucking it back again at (3), maintaining net negative energy pressure (7) inside C. The distal outflow (venous) pressure (6) enhances outflow at (4) and its elevation may turn the negative energy pressure (7) inside C into positive, increasing volume and pressure inside C chamber.
Abdalla Taha
University of Modern Science, UAE
Title: Evaluation of mRNA markers for estimating blood deposition time

Biography:
Abdalla Taha has completed his Master degree at the age of 26 years from University of Modern Science and honor his Bachelor scholarship from Khalifa University in Abu Dhabi- UAE. I’m the Sales Manager of ECHO research & development S.r,l an Italian manufacturer. I had consult and design more than ten major research labs in different fields and working with UN for design 41 vocational schools. I awarded the second place in conference competition “Forensic Science: Preparing for Next Generation” in Amity University- Dubai- UAE. Currently I am in process of publishing my thesis master thesis about Evaluation of mRNA markers for estimating blood deposition time.
Abstract:
RNA analysis provide insight into diseases, molecular identification of body fluids and mechanisms leading to death and might develop into a valuable tool for identification of the cause of death in forensic pathology. Further potential uses are the determination of the age injuries of wounds and of the post-mortem interval. In this proof-of-concept pilot study clarifies and explains principles, applications and methods by offering a comprehensive and complete overview of using mRNA markers for estimating blood deposition time which can help to evaluate the time of the crime in forensic RNA work. Study presented in this thesis aimed to estimate the time passed since blood stains found in the crime scene by calculating the time of deposited blood using particular mRNA markers and unravelling one of the principles of at what time – when during the day or night a biological evidence was left at the scene – by applying the insights from circadian biology to some open forensic cases. By analyzing 4 candidate mRNA markers expression in peripheral blood samples collected from 29 health males. The four mRNA were collected from healthy persons for the duration of the 24 hours’ day/night interval under four different groups, i.e. night/early, morning early morning/morning, morning/afternoon and afternoon/night. This study identified 2 mRNAs with statistically significant expression rhythms which are MKNK2 and PER 3. It’s found that, in general mRNA-based estimation of time categories was less accurate. The value of mRNA was demonstrated for blood deposition timing and introduced a statistical model for estimating day/night time categories based on molecular biomarkers, which shall be further validated with additional samples in the future.






















